Support Undergraduate Research in Synthetic Biology!
Bring the UO iGEM team to Paris to share their project on the world stage and support research that has potential life-saving impact!
The UO iGEM (International Genetically Engineered Machine) team is a research group of undergraduate students working in the Knight Campus for Accelerating Scientific Impact that is focused on solving real-world problems with the tools of synthetic biology. Our members come from a variety of scientific backgrounds, Biology, Chemistry, Biochemistry, Human Physiology, and even Business! Last year we designed a rapid, low-cost biosensor for detecting concussions, and this year we’re setting our sights on an engineered probiotic.
The Paris Jamboree is a scientific conference where iGEM teams from around the world collaborate with each other to advance the field of synthetic biology and network with big industry players. This year there will be more than 5000 attendees, representing 400+ teams from 66 countries. Attending the conference will allow us to present our work on the world stage, putting the University of Oregon on the map. Our research will be shown to students from the top universities in the world, as well as leading companies in the synthetic biology industry. We can join startup incubator programs to turn our proof of concept prototype into a fully fledged therapeutic, and participate in the scientific process with other iGEM teams from the rest of the world.
Attending this conference will benefit more than our students' personal development. This Jamboree will expose our members to industry and technology that is only accessible to them during this conference. Our students are ambassadors of the University of Oregon and this opportunity allows them to share their journey on an international stage. The Jamboree in Paris gives undergraduate students a chance to see high level synthetic biology that would otherwise have a barrier to entry.
Our research also has profound potential benefits in medicine. More than 14,500 people die of H. pylori every year. H. pylori infects about half of the world’s population, and this infection is the strongest risk factor for gastric cancer, the second most deadly form of cancer with a 36% 5-year survival rate. A probiotic strain effective against H. pylori can also be easily re-engineered to target any pathogen in the GI tract, making our treatment adaptable to many potential uses.
Our experimentation will cost $10,000. We need to DNA, bacteria, reagents, and consumables to make an initial design, and we need to use the results from the first design to inform our future work. With more funds, we can also work to attack other pathogens such as Salmonella enterica subsp. enterica, serovar typhi, the causative agent of typhoid fever which kills more than 200,000 people every year. Each new pathogen will likely cost $5,000 to develop a payload against.
Travel costs $3,400 per student, and we can only afford to send two of our fifteen team members with our current funds. Sending more students means that the people who worked on specific aspects of the project can explain their work and answer any questions that may come up at the conference based on their expert knowledge, helping us put our best foot forwards. A convincing presentation opens our project to support from startup incubators and can launch our research into the commercialization and mass production stage.
See below for detailed information about the project.
HGT: Hostile Gene Transfer
An engineered probiotic bacterium that eradicates H. pylori
Highlights
- UOregon iGEM’s cutting edge research on concussion biosensing brought our team to the international stage at a conference in Paris, France last year
- This year we’re setting our sights on bioengineering a living treatment for Helicobacter pylori infections
- Our treatment vehicle is a specialized probiotic strain of E. coli known as “Nissle 1917”, which has been granted GRAS (Generally Recognized as Safe) status by the FDA and is used as a living therapeutic for multiple diseases
- H. pylori is a class 1 carcinogen that infects more than half of the world’s population, and causes severe gastric diseases
- Our project addresses the urgent need for effective treatments as antibiotic resistance in H. pylori and other bacteria continues to rise.
- The current state of the art antibiotic/drug cocktail fails in 10-30% of patients
- By engineering a genetically modified bacterium, we are paving the way for a safe and sustainable alternative to combat bacterial infections in the gastrointestinal tract.
- The end goal of this project is a probiotic that can be stored in a shelf-stable pill and deployed at a comparable cost to other E. coli nissle based therapeutics of approximately $1 per dose
Our Motivation
Helicobacter pylori (H. pylori) is a gram negative bacterial pathogen that, as of 2015, resided in the gastrointestinal tract of an estimated 50% of the world’s population1, largely affecting developing countries and communities. Fortunately, most of these infections are asymptomatic, meaning that the bacterium stays in the stomach and the infected don’t feel any different. The triggers that switch a benign H. pylori infection to a pathogenic one aren’t entirely understood, but studies have found associations with changes in diet, smoking, and even stress.
When H. pylori switches to its virulent phase, it tunnels through the stomach’s natural defensive mucous layer in a corkscrewing motion and starts to attack sensitive epithelial cells directly. Once H. pylori reaches the epithelial layer, it begins to inject toxins into stem cells, which can cause them to replicate uncontrollably, a signature feature of malignant cancers. It has been classified as a class 1 carcinogen by the International Agency for Research in Cancer of the World Health Organization, the same category as asbestos and the nuclear waste product strontium-902,3. It is the cause of the majority of cases of gastric adenocarcinoma (the most common type of stomach cancer), and also causes low-grade B-cell gastric lymphomas (MALTomas)4,5. H. pylori is the causative agent for up to 95% of duodenal ulcers and 70–80% of gastric ulcers6.
The human immune system is significantly less effective GI tract than in the rest of the body, and is especially low in the stomach (H. pylori’s habitat), due to both extreme acidity and mucosal dynamics7. H. pylori’s natural defense systems are too good for our immune system to fight, and patients cannot eradicate H. pylori infection without therapeutic support. The best tools we have today to fight H. pylori are broad spectrum antibiotics, which have become significantly less effective in recent years8. Current antibiotic-only treatments are only somewhat effective, varying widely depending on drug choice with a pooled eradication rate of 79%9.
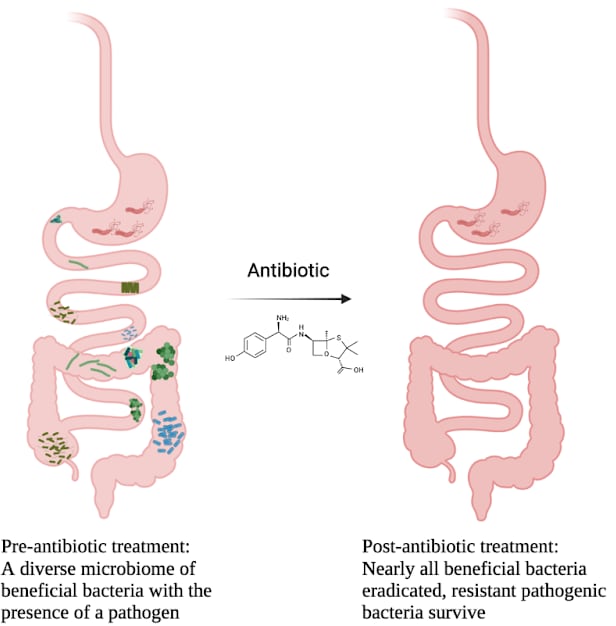 Infographic showing the indiscriminate effects of broad-spectrum antibiotics
Infographic showing the indiscriminate effects of broad-spectrum antibioticsThis number will only become lower as H. pylori evolves to fight more and more antibiotics. It has already developed resistance to nalidixic acid, trimethoprim, vancomycin, and most sulfonamide compounds. Worse still now we’re discovering isolates that are becoming resistant to metronidazole (a component of FDA-approved H. pylori therapy) and clarithromycin10. We are running out of antibiotics that work to fight H. pylori and other bacterial infections are beginning to become resistant to even stores of “antibiotics of last resort”11.
Our Project
Using our team’s expertise in genetic engineering and the support of the global iGEM network, we will develop a genetically modified bacterium to seek and destroy H. pylori in the gut without using any antibiotics. We’ll be working with “Nissle 1917” a probiotic strain of E. coli that’s already in use in the US for treating certain GI conditions and is sold under the brand name Mutaflor®. Since this bacterium has already been granted Generally Recognized as Safe (GRAS) status by the FDA, our path to deployment is significantly easier than other groups which start from scratch and need to secure their own FDA approvals.
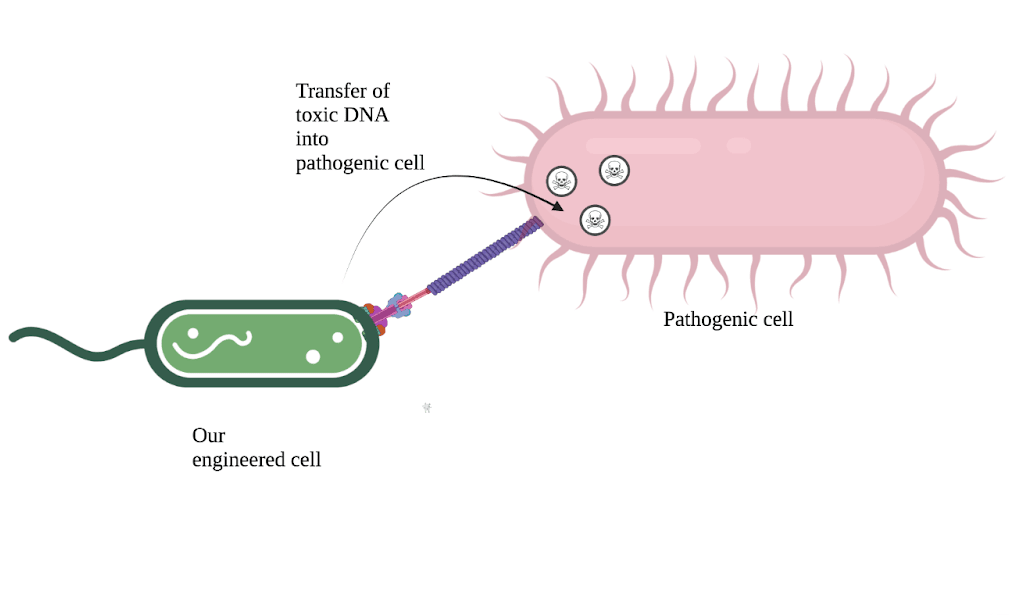 Diagram of our bacterial conjugation system; our engineered cell delivers toxic DNA into a pathogenic cell for species-specific antimicrobial activity
Diagram of our bacterial conjugation system; our engineered cell delivers toxic DNA into a pathogenic cell for species-specific antimicrobial activityOur engineered strain will specifically recognize H. pylori cells using protein binders we design ourselves in the lab, and deliver a malicious DNA payload through a process known as “horizontal gene transfer”. Once injected, this DNA will use H. pylori’s own molecular machinery to disrupt the biofilm-forming colonies, spread itself to dormant H. pylori cells, and eventually trigger the injected cell to kill itself. This is the same principle as cutting edge “phage therapies”, but using bacteria allows us to deploy our treatment at a significantly lower cost while having a greater control over the specificity of our attacks.
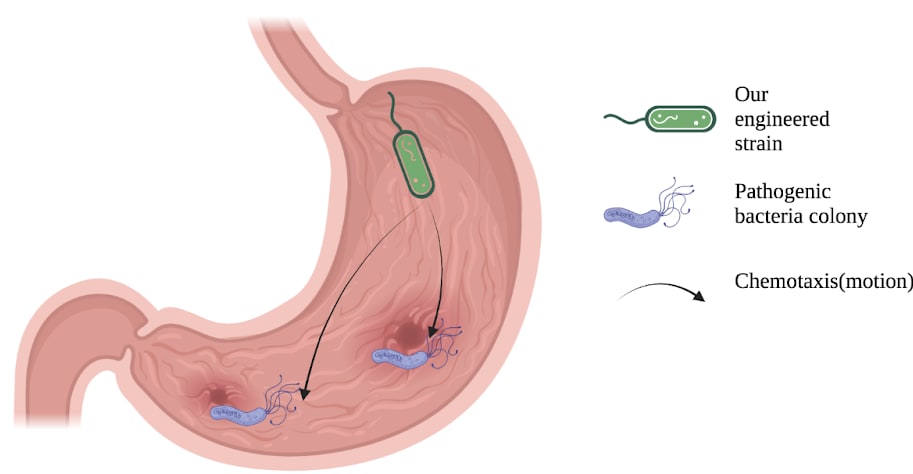 Diagram of bacterial chemotaxis, our cells are following a chemical gradient that leads them towards our pathogen of interest
Diagram of bacterial chemotaxis, our cells are following a chemical gradient that leads them towards our pathogen of interestWe are taking a uniquely modular approach that will allow future work to modify our platform to be targeted towards nearly any pathogenic GI tract bacteria, and allows us to develop these parts in parallel. Other common pathogen examples, such as Salmonella typhi (responsible for typhoid fever), enterotoxigenic Escherichia coli, Salmonella enterica, Clostridioides difficile could also be targeted using our system.
Once commercialized, our system could be targeted towards these or any other pathogenic gut bacterium. Bacteria in our gut outnumber our cells 10 to 1, and every person’s gut microbiome is slightly different. It’s impossible to say how many of these bacteria could turn into a bacterial infection, but we could build and deploy a new version of our strain in months to target that species directly.
- We are computationally designing De Novo binders for unique surface features of H. pylori. These binders will determine the specificity of our conjugation machinery, and allow us to deliver our genetic payload exclusively into target cells. Our computational protocols can be trivially adapted to target surface features of any well characterized pathogen.
- We plan to create an E. coli biofilm in the stomach with functionalized CsgA monomers. These functionalities will include an H. pylori urease inhibitor, H. pylori surface binders, and a ferritin domain for magnetotaxis. These functionalities could also be replaced with ones more relevant to other pathogenic bacteria that favor biofilm formation in different locations of the GI tract.
- We are exploring chemotaxis pathways to move towards H. pylori cells, moving past the mucosal layers of the stomach and approaching epithelial cells. These pathways could be replaced with any that follow a relevant chemical gradient, or trail left by any target bacteria.
By engineering a variety of biological systems for stomach colonization, navigation to H. pylori, and conjugation to deliver a fatal DNA payload, we are able to create an E. coli-based therapeutic that is specific, modular, and safe. Our E. coli-based solution does not rely on the use of antibiotics, which can be harmful when used repeatedly in high doses, and are becoming increasingly ineffective against bacterial infections.
We truly believe that every little step we take today can make a substantial difference in the lives of countless individuals in the future. It is with utmost sincerity and gratitude that we seek your benevolent support to make our participation in this convention a reality. Your commitment to empowering the next generation of researchers and change-makers will undoubtedly leave an indelible mark on our team, university, and the world at large.
References:
(1) Hooi, J. K. Y.; Lai, W. Y.; Ng, W. K.; Suen, M. M. Y.; Underwood, F. E.; Tanyingoh, D.; Malfertheiner, P.; Graham, D. Y.; Wong, V. W. S.; Wu, J. C. Y.; Chan, F. K. L.; Sung, J. J. Y.; Kaplan, G. G.; Ng, S. C. Global Prevalence of Helicobacter Pylori Infection: Systematic Review and Meta-Analysis. Gastroenterology 2017, 153 (2), 420–429. https://doi.org/10.1053/j.gastro.2017.04.022.
(2) A Review of Human Carcinogens; Centre international de recherche sur le cancer, Ed.; IARC monographs on the evaluation of carcinogenic risks to humans; International agency for research on cancer: Lyon, 2012.
(3) Schistosomes, Liver Flukes and Helicobacter Pylori: This Publication Represents the Views and Expert Opinions of an IARC Working Group on the Evaluation of Carcinogenic Risks to Humans, Which Met in Lyon, 7 - 14 June 1994; International Agency for Research on Cancer, Ed.; IARC monographs on the evaluation of carcinogenic risks to humans; IARC: Lyon, 1994.
(4) Correa, P.; Malcom, G.; Schmidt, B.; Fontham, E.; Ruiz, B.; Bravo, J. C.; Bravo, L. E.; Zarama, G.; Realpe, J. L. Review Article: Antioxidant Micronutrients and Gastric Cancer. Aliment Pharmacol Ther 1998, 12 Suppl 1, 73–82. https://doi.org/10.1111/j.1365-2036.1998.00006.x.
(5) Isaacson, P. G. Recent Developments in Our Understanding of Gastric Lymphomas. Am J Surg Pathol 1996, 20 Suppl 1, S1-7. https://doi.org/10.1097/00000478-199600001-00002.
(6) Dunn, B.; Cohen, H.; Blaser, M. H. Pylori. Microbiol. Rev 1997, 10, 720–741.
(7) Cieza, R. J.; Cao, A. T.; Cong, Y.; Torres, A. G. Immunomodulation for Gastrointestinal Infections. Expert Rev Anti Infect Ther 2012, 10 (3), 391–400. https://doi.org/10.1586/eri.11.176.
(8) Ventola, C. L. The Antibiotic Resistance Crisis. P T 2015, 40 (4), 277–283.
(9) Effectiveness of eradication therapy for Helicobacter pylori infection in Africa: a systematic review and meta-analysis | BMC Gastroenterology | Full Text. https://bmcgastroenterol.biomedcentral.com/articles/10.1186/s12876-023-02707-5 (accessed 2023-06-29).
(10) On, S. L. W.; Lee, A.; O’Rourke, J. L.; Dewhirst, F. E.; Paster, B. J.; Fox, J. G.; Vandamme, P. Helicobacter. In Bergey’s Manual of Systematics of Archaea and Bacteria; John Wiley & Sons, Ltd, 2015; pp 1–1. https://doi.org/10.1002/9781118960608.gbm01073.
(11) McKenna, M. Antibiotic Resistance: The Last Resort. Nature 2013, 499 (7459), 394–396. https://doi.org/10.1038/499394a.
$50
General Lab Items
This donation helps us to purchase lab items such as pipette tips, petri dishes, and test tubes!
$100
Lab Funds
$100 buys one length of DNA or an enzyme, we need these to conduct our research! We will send donors a specialized thank you gift created by us.
$3,000
Send one student to Paris!
$3,000 will cover flight, housing, transportation, and the Grand Jamboree ticket for one student. We will acknowledge your donation with your name or organization on our t-shirt and in our materials for Paris in addition to a thank you gift designed by our team.
$5,000
Platinum Sponsor
Donations above $5,000 will be acknowledged on our t-shirts and written materials and sent a specialized thank you gift created by our team.




